Information about the research group
Leader
Gergely Róna, PhD
General research interests
Our laboratory investigates how cells maintain their genomic integrity. By using genetic, biochemical, and proteomic approaches, we unravel molecular mechanisms to gain a better understanding of diseases. We work towards providing the foundation for innovative, novel therapeutic approaches.
Overview of what we do
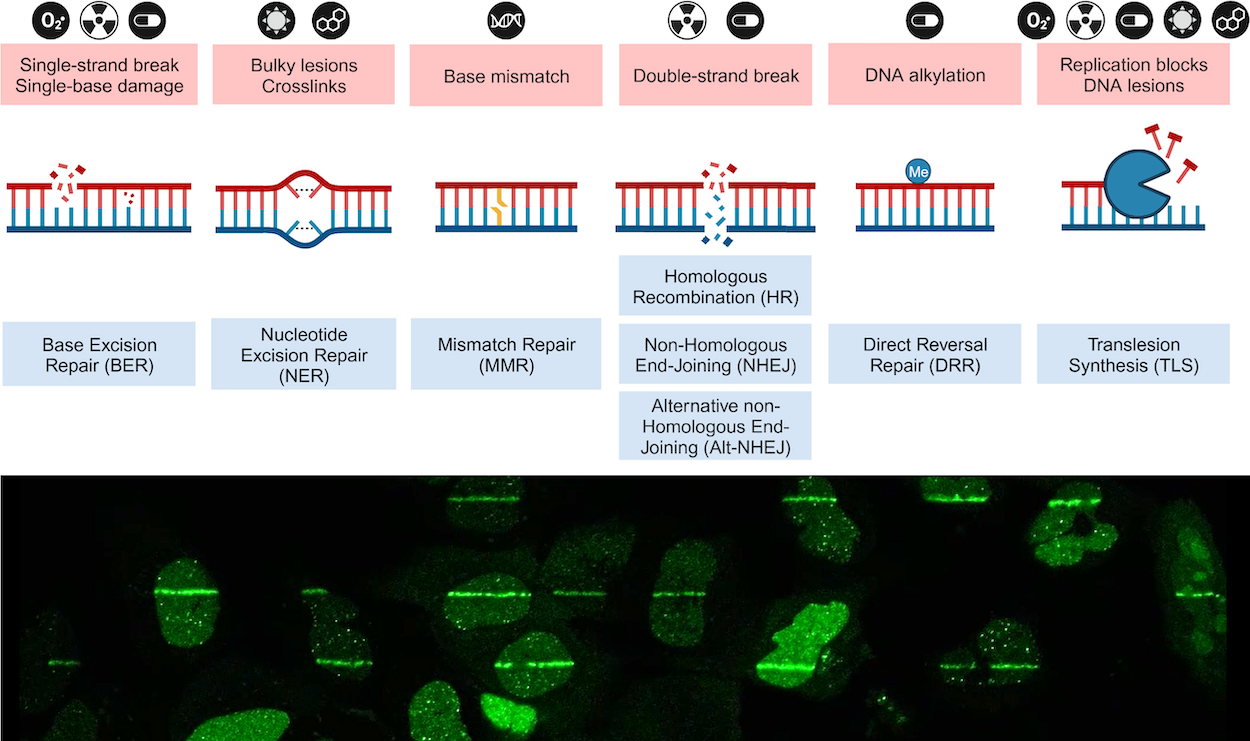
Our laboratory seeks to gain a better understanding of the fundamental processes that maintain genomic integrity within our cells. It is remarkable how information encoded in our genome is maintained with unparalleled precision, yet at the same time is flexible enough to be able to adopt to environmental changes. DNA repair processes are crucial aspects of biology as mis-regulation of these mechanisms are often the driving force behind several types of cancer and neurodegenerative diseases. By unraveling the underlying molecular mechanisms of these diseases, we aim to provide the foundation of novel therapeutic approaches. In our work we rely on genetic, biochemical, and proteomic approaches in mammalian cells as well as collaborations with structural biologists and mouse geneticists.
Neuronal DNA repair
Molecular mechanisms of neuronal DNA repair. Neurological disorders are one of the leading cause of disability and death worldwide. Our goal is to obtain a better understanding of defects in neuronal DNA repair, and to exploit this knowledge to identify new vulnerabilities in neurodegeneration. The nervous system is highly susceptible to all forms of stress, including DNA damage, due to its large energy requirements, high transcriptional activity, and the long lifespan of these cells. Data suggests that mutations in DNA repair genes are associated with several neurological abnormalities and somatic tri/dinucleotide expansions, which plays a critical role in the development of neurodegenerative diseases such as Huntington’s disease (HD), or Amyotrophic lateral sclerosis (ALS). Neurons can repair and protect their genome against DNA damage under physiological conditions despite lacking DNA replication, which limits their opportunity for DNA repair. What remains in question, however, are the underlying molecular mechanisms behind these repair processes and how these non-renewable cells maintain their genomic integrity over many decades of life.
Nuclease inhibition in therapeutics
Inhibitor screening and design. Our goal is to identify small molecule inhibitors against relevant exonucleases to aid existing therapies in oncology. Using a novel, high-throughput activity assays, our aim is to identify small molecule inhibitors for exonucleases that are relevant in the immunotherapy field. Our efforts will aid both radiation and chemotherapeutics treatments to promote IFN-dependent antitumor immunity.
Post-Replicative DNA Damage Bypass
Functions of translesion DNA polymerases. Each of our cells must cope with tens of thousands of DNA lesions a day. Depending on the exact nature of these lesions and the cell cycle phase the cells are in, the repair will be carried out by one of the DNA repair pathways. Cells face a special challenge when they encounter a DNA lesion when either performing DNA replication in S phase or while performing DNA repair synthesis (which is the gap-filling step of the DNA repair process). The catalytic site of the replicative DNA polymerases, utilized during the DNA elongation processes, is compact and cannot accommodate most DNA lesions. Consequently, DNA synthesis arrests at most forms of DNA lesions. This is when a polymerase switch is initialized on PCNA, where the replicative DNA polymerase is replaced by translesion polymerases to bypass these lesions. This process is called translesion synthesis (TLS). While TLS polymerase activation is well studied during S phase in replicating cells, almost nothing is known about their function in other cell cycle phases (like G0 and G1) or in non-replicating cells.
Main publications
Zhang, Q., Kerzhnerman, M. A., García-Vázquez, N., Rona, G. Visualizing Single-Stranded DNA Foci in the G1 Phase of the Cell Cycle. J. Vis. Exp (2023).
Rona, G., Zeke, A., Miwatani-Minter, B., de Vries, M., Kaur, R., Schinlever, A., Garcia, S. F., Goldberg, H. V., Wang, H., Hinds, T. R., Bailly, F., Zheng, N., Cotelle, P., Desmaele, D., Landau, N. R., Dittmann, M. & Pagano, M. The NSP14/NSP10 RNA repair complex as a Pan-coronavirus therapeutic target. Cell Death Differ 29, 285-292 (2022).
Simoneschi, D., Rona, G., Zhou, N., Jeong, Y. T., Jiang, S., Milletti, G., Arbini, A. A., O’Sullivan, A., Wang, A. A., Nithikasem, S., Keegan, S., Siu, Y., Cianfanelli, V., Maiani, E., Nazio, F., Cecconi, F., Boccalatte, F., Fenyo, D., Jones, D. R., Busino, L. & Pagano, M. CRL4(AMBRA1) is a master regulator of D-type cyclins. Nature 592, 789-793 (2021).
Miwatani-Minter, B. & Rona, G. Laser Micro-Irradiation to Study DNA Recruitment During S Phase. J Vis Exp (2021).
Maiani, E., Milletti, G., Nazio, F., Holdgaard, S. G., Bartkova, J., Rizza, S., Cianfanelli, V., Lorente, M., Simoneschi, D., Di Marco, M., D’Acunzo, P., Di Leo, L., Rasmussen, R., Montagna, C., Raciti, M., De Stefanis, C., Gabicagogeascoa, E., Rona, G., Salvador, N., Pupo, E., Merchut-Maya, J. M., Daniel, C. J., Carinci, M., Cesarini, V., O’Sullivan, A., Jeong, Y. T., Bordi, M., Russo, F., Campello, S., Gallo, A., Filomeni, G., Lanzetti, L., Sears, R. C., Hamerlik, P., Bartolazzi, A., Hynds, R. E., Pearce, D. R., Swanton, C., Pagano, M., Velasco, G., Papaleo, E., De Zio, D., Maya-Mendoza, A., Locatelli, F., Bartek, J. & Cecconi, F. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 592, 799-803 (2021).
Palinkas, H. L., Bekesi, A., Rona, G., Pongor, L., Papp, G., Tihanyi, G., Holub, E., Poti, A., Gemma, C., Ali, S., Morten, M. J., Rothenberg, E., Pagano, M., Szuts, D., Gyorffy, B. & Vertessy, B. G. Genome-wide alterations of uracil distribution patterns in human DNA upon chemotherapeutic treatments. Elife 9 (2020).
Rona, G., Roberti, D., Yin, Y., Pagan, J. K., Homer, H., Sassani, E., Zeke, A., Busino, L., Rothenberg, E. & Pagano, M. PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading. Elife 7 (2018).
Rona, G., Scheer, I., Nagy, K., Palinkas, H. L., Tihanyi, G., Borsos, M., Bekesi, A. & Vertessy, B. G. Detection of uracil within DNA using a sensitive labeling method for in vitro and cellular applications. Nucleic Acids Res 44, e28 (2016).
Dankert, J. F., Rona, G., Clijsters, L., Geter, P., Skaar, J. R., Bermudez-Hernandez, K., Sassani, E., Fenyo, D., Ueberheide, B., Schneider, R. & Pagano, M. Cyclin F-Mediated Degradation of SLBP Limits H2A.X Accumulation and Apoptosis upon Genotoxic Stress in G2. Mol Cell 64, 507-519 (2016).
Rona, G., Borsos, M., Ellis, J. J., Mehdi, A. M., Christie, M., Kornyei, Z., Neubrandt, M., Toth, J., Bozoky, Z., Buday, L., Madarasz, E., Boden, M., Kobe, B. & Vertessy, B. G. Dynamics of re-constitution of the human nuclear proteome after cell division is regulated by NLS-adjacent phosphorylation. Cell Cycle 13, 3551-3564 (2014).
Muha, V., Horvath, A., Bekesi, A., Pukancsik, M., Hodoscsek, B., Merenyi, G., Rona, G., Batki, J., Kiss, I., Jankovics, F., Vilmos, P., Erdelyi, M. & Vertessy, B. G. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet 8, e1002738 (2012).
Current collaborations:
Anna Pluciennik, Thomas Jefferson University, USA
Beata Vertessy, HUN-REN TTK, Hungary
Beatrix Ueberheide, New York University School of Medicine, USA
Bostjan Kobe, University of Queensland, Australia
David Fenyö, NYU Grossman School of Medicine, USA
Didier Desmaële, Institut Galien, Université Paris-Saclay, France
Eli Rothenberg, NYU Grossman School of Medicine, USA
Lorinc S. Pongor, Hungarian Centre of Excellence for Molecular Medicine, University of Szeged, Hungary
Manor Askenazi, Biomedical Hosting LLC, USA
Meike Dittmann, NYU Grossman School of Medicine, USA
Michele Pagano, NYU Grossman School of Medicine / HHMI, USA
Nathaniel R. Landau, NYU Grossman School of Medicine, USA
Ning Zheng, University of Washington / HHMI, USA
Philippe Cotelle, University of Lille, INSERM, France
Piotr Sicinski, Dana-Farber Cancer Institute, USA
Zachary D. Nagel, Harvard T.H. Chan School of Public Health, USA
Group photo (2024)