Leader
Dr. Sándor Spisák (publications)
General research interest
Our research is centered around investigating the functional role of the non-coding genome in cancer development using colon cancer models. By employing epigenetic analysis, genome editing techniques, and computational approaches, we aim to unravel the functional significance of non-coding genetic variants in disease susceptibility. Additionally, we seek to identify and characterize functional regulatory elements that contribute to disease development. Through these efforts, our ultimate objective is to identify therapeutic vulnerabilities and leverage this knowledge for early disease detection.
Main research topics
- Germline risk variants
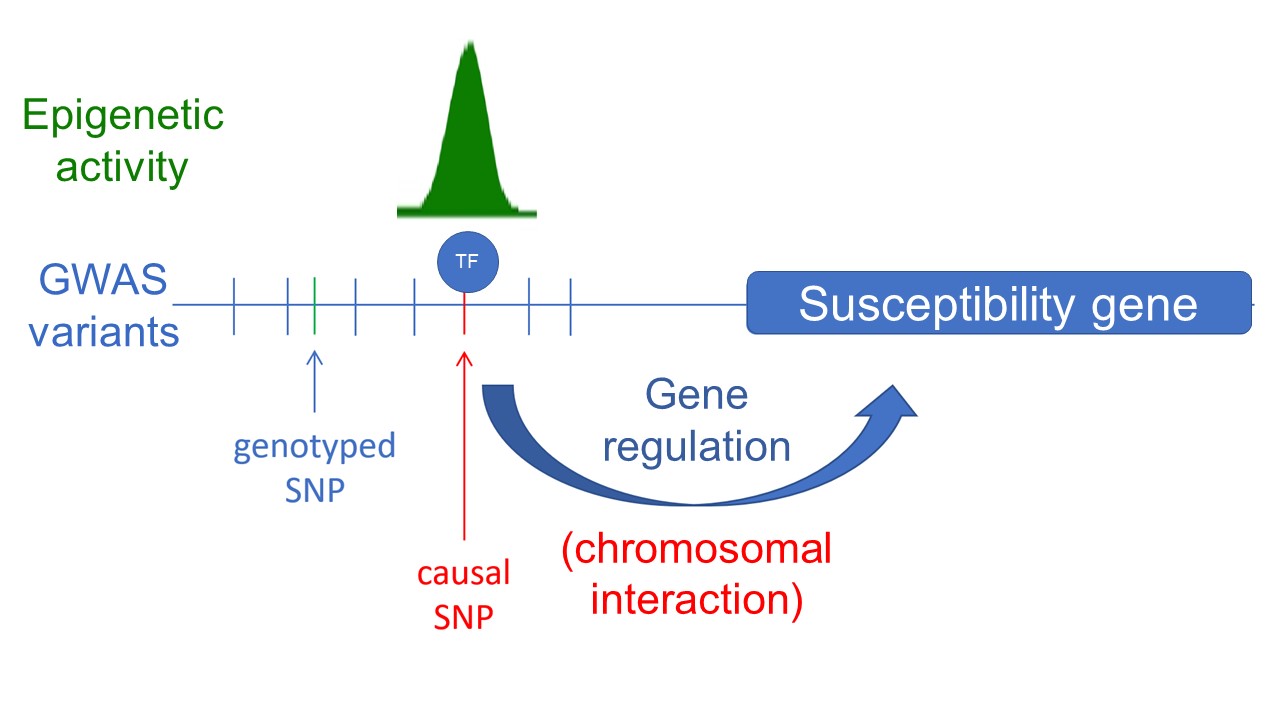
We are dedicated to unraveling the role and impact of inherited non-coding mutations in cancer development, primarily identified through genome-wide association studies (GWAS). However, studying their influence on cancer risk poses significant challenges, including genetic correlation (linkage disequilibrium), cis-regulation (no genetic code), mild or weak phenotypes, complex genetic variants, limited model systems and methodological constraints. To overcome these hurdles, our primary objective is to identify and comprehensively investigate causal variants associated with cancer development. We employ precise genome and epigenome editing techniques, single-cell cloning, and cell-based assays to functionally annotate these causal variants and establish their connection to disease susceptibility genes (Spisak et al, Nat. Med, 2015 link). Through these approaches, we aim to deepen our understanding of their functional characteristics and their contribution to disease susceptibility. Furthermore, we strive to develop more sensitive systems and assays that leverage sophisticated phenotypes such as gene expression, cellular states, and morphology, with the ultimate goal of achieving scalability in our research endeavors.
- Enhancer biology
Cis-regulatory elements, known as enhancers, are specific segments of the non-coding DNA that exert remote control over distant gene transcription. These elements interact with transcription factors (TFs), which are trans-acting proteins, to enhance the expression of target genes located on the same DNA molecule. Several large-scale genome-wide sequencing initiatives have provided evidence that enhancers are frequently transcribed into long non-coding RNA (lncRNA) or enhancer RNA (eRNA). The levels of these transcripts often exhibit a correlation with the expression levels of the target gene’s mRNA. Our research aims to identify cancer-specific enhancers and investigate their mechanisms of action, including TF binding, activation, interaction, and tissue-specific operation. Additionally, we seek to understand the consequences of altered target gene expression associated with cancer development. (Takeda and Spisak et al, Cell, 2018 link)
- Early cancer detection
Epigenetic alterations, particularly DNA methylation and histone modifications, are known to exhibit tissue-specific and cancer-specific patterns. Investigating the rearrangement of the epigenetic landscape during cancer development, coupled with the utilization of liquid biopsy technology, holds great potential for advancing early cancer detection strategies. By integrating these approaches, we aim to enhance our understanding of epigenetic changes in cancer and develop effective methods for early detection. (Nuzzo, Berchuck, Korthauer and Spisak et al, Nat.Med. 2020 link)
Publications(2023)
Spisak S, Tisza V, Nuzzo PV, Seo JH, Pataki B, Ribli D, Sztupinszki Z, Bell C, Rohanizadegan M, Stillman DR, Alaiwi SA, Bartels AB, Papp M, Shetty A, Abbasi F, Lin X, Lawrenson K, Gayther SA, Pomerantz M, Baca S, Solymosi N, Csabai I, Szallasi Z, Gusev A, Freedman ML. A biallelic multiple nucleotide length polymorphism explains functional causality at 5p15.33 prostate cancer risk locus. Nat Commun. 2023 Aug 23;14(1):5118. doi: 10.1038/s41467-023-40616-z. link
Vízkeleti L., Spisák S., Rewired Metabolism Caused by the Oncogenic Deregulation of MYC as an Attractive Therapeutic Target in Cancers, Cells 2023, 12(13), 1745; link
Szmolka A, Gellért Á, Szemerits D, Rapcsák F, Spisák S, Adorján A. Emergence and Genomic Features of a mcr-1 Escherichia coli from Duck in Hungary. Antibiotics (Basel). 2023 Oct 7;12(10):1519. doi: 10.3390/antibiotics12101519. PMID: 37887221 link
Sahgal P, Patil DT, Bala P, Sztupinszki ZM, Tisza V, Spisak S, Luong AG, Huffman B, Prosz A, Singh H, Lazaro JB, Szallasi Z, Cleary JM, Sethi NS. Replicative stress in gastroesophageal cancer is associated with chromosomal instability and sensitivity to DNA damage response inhibitors. iScience. 2023 Oct 12;26(11):108169. doi: 10.1016/j.isci.2023.108169. eCollection 2023 Nov 17. PMID: 37965133 link
Prosz A, Duan H, Tisza V, Sahgal P, Topka S, Klus GT, Börcsök J, Sztupinszki Z, Hanlon T, Diossy M, Vizkeleti L, Stormoen DR, Csabai I, Pappot H, Vijai J, Offit K, Ried T, Sethi N, Mouw KW, Spisak S, Pathania S, Szallasi Z. Nucleotide excision repair deficiency is a targetable therapeutic vulnerability in clear cell renal cell carcinoma. Sci Rep. 2023 Nov 23;13(1):20567. doi: 10.1038/s41598-023-47946-4. link
Prosz A., Pipek O., Börcsök J., Palla G., Szallasi Z., Spisak S.*, Csabai I. Biologically informed deep learning for explainable epigenetic clocks. Sci Rep. 2023 Dec 21
Pre-prints
Vizkeleti L., Kiss C., Tisza V., Szigeti A., Gellert A., Csabai I., Pongor LS., Spisak S.,# (2023) Epigenetic regulation explains the functionality behind colon cancer specific biomarker Septin9. BioRxiv
Gusev A.*, Spisak S.*, Fay AP., Carol H., Vavra KC., Signoretti S., Tisza V., Pomerantz M., Abbasi F., Seo JH., Choueiri TK., Lawrenson K., Freedman ML., Allelic imbalance reveals widespread germline-somatic regulatory differences and prioritizes risk loci in Renal Cell Carcinoma. BioRxiv
Spisak S., Chen D., Likasitwatanakul P., Doan P., Li X., Vizkeleti L., Tisza V., Silva PD., Giannakis M., Wolpin B., Qi J., Sethi NS., Utilizing a dual endogenous reporter system to identify functional regulators of aberrant stem cell and differentiation activity in colorectal cancer. BioRxiv
Group photo (2023)

Collaborations
International
Dana-Farber Cancer Institute, Boston, MA, USA
Boston Children`s Hospital, Boston, MA, USA
University of Massachusetts, Boston, MA, USA
Weill Cornell Medicine, New York, NY, USA
Medical University of Vienna, Vienna, Austria
Paris-Lodron-University Salzburg, Salzburg, Austria
Institutional
HUN-REN Biological Research Centre, Szeged
HUN-REN Research Centre for Natural Sciences, Institute of Organic Chemistry, Budapest
HUN-REN Centre for Agricultural Research, Budapest
National
Semmelweis University, Budapest
Eotvos Lorand University, Budapest
National Korányi Institute for Pulmonology, Budapest
Hungarian Centre Of Excellence For Molecular Medicine (HCEMM), Szeged